MISSION
Microbial communities are the engines that drive the biosphere, playing critical biochemical roles in ecosystems and hosts, from soils to oceans to the human gut. What rules govern how communities are put together (assembly) and what chemistry they’re doing (function)?
The Gowda Lab studies how microbial nitrogen cycling communities work. We take a systems biology approach, emphasizing a quantitative, dynamical understanding of how molecular, physiological, and ecological pieces of a system give rise to the whole. Ongoing projects and areas of interest include the resource allocation biology of bacterial denitrification, nitrogen cycling in biofilms, nutrient limited growth, and novel techniques for profiling gene expression in consortia. We are an interdisciplinary team of researchers in the Department of Microbiology and Biophysics Graduate Program at the Ohio State University.
NEWS
- Aug 2025: Big congrats to Bryce Guidry for successfully completing his PhD candidacy examinations in the Biophysics Graduate Program!
- May 2025: We welcome summer REU student Emily Yoon to the lab! Emily is a rising junior at Williams College, and will be working on developing a new model system for studying the community ecology of denitrification.
- April 2025: Undergraduate researcher Aouss Azzouz won another poster award, this time at the Emerging Researchers National Conference in STEM. Also very proud to announce that he will be starting med school at UPenn next Fall. Congrats Aouss!
- February 2025: We are thrilled to have Dominic Cipiti joining the lab as a Microbiology PhD student. He will take a resource-allocation perspective to the study of denitrification physiology.
- January 2025: We welcome Bryce Guidry and Anja Steinert to the lab! Bryce joins as a PhD student through the Biophysics Graduate Program and will be working on single-cell approaches for studying bacteria. Anja will be taking over as lab manager.
- November 2024: Summer REU student Aouss Azzouz won a poster award for presenting his work in the Gowda Lab at ABRCMS! Congrats Aouss!
- July 2024: Our paper on how interactions shape denitrifying communities in the soil microbiome is now out in Nature Microbiology.
PEOPLE
Karna Gowda, PhD [CV]
Principal InvestigatorDepartment of Microbiology
Biophysics Graduate Program
The Ohio State University

Dominic Cipiti
PhD Student, MicrobiologyBS in Biology, Baldwin Wallace University
Ava Galek
Undergraduate Research Assistant
Bryce Guidry
PhD Candidate, BiophysicsBS in Biophysics, Emory University
Aayushi Shah
High School InternAnja Steinert
Lab ManagerBS in Biology, The Ohio State University

Kuromatsu
DogAlumni
- Emily Yoon (Williams College), SROP Researcher (2025)
- Molly Easton, Undergraduate Lab Assistant (2024-2025)
- Aouss Azzouz (Earlham College→UPenn Medical School), SROP Researcher (2024)
PUBLICATIONS
Recent highlights

An old truism in microbial ecology is that "everything is everywhere but the environment selects". But we know that interactions between organisms in microbial communities are widespread—shouldn't that matter as well? In this paper, we show that community assembly is shaped by microbial physiology and interactions, not just environmental factors. Using a statistical approach to analyze global topsoil microbiome data, we uncovered a novel link between denitrification reductase gene abundances and soil pH. Lab experiments and isolate characterizations showed that low pH promotes toxin-antitoxin codependency between strains, explaining statistical patterns observed in soils around the planet.
News & Views by Daniel Amor
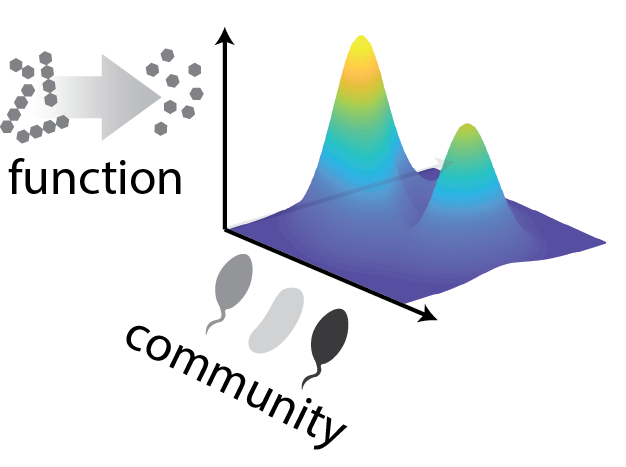
Imagine opening a freezer box full of strains and asking which combination of them would be best at performing a given function (e.g., producing or degrading a compound of interest). Screening all combinations of 10 strains requires at least a thousand experiments, and all combinations of 20 strains requires a million. Here, we show that searching the space of community configurations may in fact be much easier than brute force. We do this by applying statistical learning to the concept of community-function landscapes, showing in datasets and models that these landscapes are often smooth and thus possible to explore with only a handful of experiments.
Dispatch by Avi Flamholz and Dianne Newman
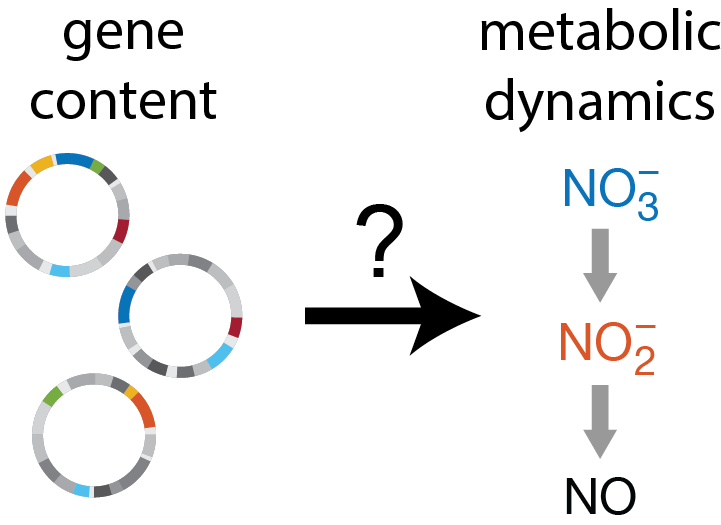
In natural microbial communities, measuring gene content is one of the easiest things to do, while measuring metabolic process rates is one of the hardest. But what if we could predict the latter from the former? In this paper, we establish a proof of concept that this is possible using bacterial denitrification as a model system. Through experimental measurements of synthetic communities composed of diverse isolates, we use statistical learning to map genotypes to phenotypes, and consumer-resource modeling to map phenotypes to community metabolic dynamics.
2025
Urvoy, M., Howard-Varona, C., Owusu-Ansah, C., Stai, A. J., Bouranis, J. A., Burris, M., Solonenko, N., Hettich, R. L., Holmfeldt, K., Tfaily, M., Gowda, K., Sullivan & M. B. (2025). Phage resistance mutations in a marine bacterium impact biogeochemically relevant cellular processes. Nature Microbiology, 11, 195–210. doi: 10.1038/s41564-025-02202-5
2024
Crocker, K., Lee, K. K., Chakraverti-Wuerthwein, M., Li, Z., Tikhonov, M., Mani, M., Gowda, K., & Kuehn, S. (2024). Environmentally dependent interactions shape patterns in gene content across natural microbiomes. Nature Microbiology, 9(8), 2022–2037. doi: 10.1038/s41564-024-01752-4
2023
Skwara, A., Gowda, K., Yousef, M., Diaz-Colunga, J., Raman, A. S., Sanchez, A., Tikhonov, M., & Kuehn, S. (2023). Statistically learning the functional landscape of microbial communities. Nature Ecology & Evolution, 7(11), 1823–1833. doi: 10.1038/s41559-023-02197-4
Diaz-Colunga, J., Skwara, A., Gowda, K., Diaz-Uriarte, R., Tikhonov, M., Bajic, D., & Sanchez, A. (2023). Global epistasis on fitness landscapes. Philosophical Transactions of the Royal Society B, 378(1877), 1–20. doi: 10.1098/rstb.2022.0053
2022
Gowda, K., & Kuehn, S. (2022). Microbial biofilms: An ecological tale of Jekyll and Hyde. Current Biology, 32(24), R1349–R1351. doi: 10.1016/j.cub.2022.10.068
Gowda, K., Ping, D., Mani, M., & Kuehn, S. (2022). Genomic structure predicts metabolite dynamics in microbial communities. Cell, 185(3), 530-546.e25. doi: 10.1016/j.cell.2021.12.036
Gopalakrishnappa, C., Gowda, K., Prabhakara, K. H., & Kuehn, S. (2022). An ensemble approach to the structure-function problem in microbial communities. iScience, 25(2), 103761. doi: 10.1016/j.isci.2022.103761
2020
Fraebel, D. T., Gowda, K., Mani, M., & Kuehn, S. (2020). Evolution of Generalists by Phenotypic Plasticity. iScience, 23(11), 1–22. doi: 10.1016/j.isci.2020.101678
PhD
Gandhi, P., Werner, L., Iams, S., Gowda, K., & Silber, M. (2018). A topographic mechanism for arcing of dryland vegetation bands. Journal of the Royal Society Interface, 15(147). doi: 10.1098/rsif.2018.0508
Gowda, K., Iams, S., & Silber, M. (2018). Signatures of human impact on self-organized vegetation in the Horn of Africa. Scientific Reports, 8(1), 1–8. doi: 10.1038/s41598-018-22075-5
Gowda, K., Chen, Y., Iams, S., & Silber, M. (2016). Assessing the robustness of spatial pattern sequences in a dryland vegetation model. Proceedings of the Royal Society A, 472(2187). doi: 10.1098/rspa.2015.0893
Gowda, K., & Kuehn, C. (2015). Early-warning signs for pattern-formation in stochastic partial differential equations. Communications in Nonlinear Science and Numerical Simulation, 22(1–3), 55–69. doi: 10.1016/j.cnsns.2014.09.019
Gowda, K., Riecke, H., & Silber, M. (2014). Transitions between patterned states in vegetation models for semiarid ecosystems. Physical Review E, 89(2), 1–8. doi: 10.1103/PhysRevE.89.022701